Table 1. Preliminary counts of disomy in Mink sperm cells for chromosome 8 and 9 and 11
. Incidence of disomy in mink sperm by strong centromere
repeat probes for the chromosome 2, 5, 8, 9, 11 and Y.
Division of Animal Genetics, Department of Animal Science and Animal Health, The Royal Veterinary and Agricultural University, Bülowsvej 13, DK-1870 Frederiksberg C, Denmark
and
Klaus Bruusgaard
Department of Biochemical and Molecular Genetics,
Odense University Hospital, Sdr. Bouleward 29, DK-5000 Odense C
Abstract
Strong repeat probes have been developed for 6 mink chromosomes. They can be utilized to estimate the rate of disomy in sperm cells by means of in flourescent in situ hybridization (FISH). This can initially give an estimate of the non disjunction rate in normal males and it can be developed for studying animals with fertility problems. The disomy rate has preliminary been estimated to 0,51, 0,54 and 0,27 per cent for chromosome 8, 9 and 11, respectively.
Key words: American Mink, sperm, disomy, centromere repeat probes
Introduction
The America mink (Mustela vison) belongs to the order of Carnivora, and the economic aspects
of this animal is indicated by the extensive mink fur production in the Scandinavian countries,
USA, Canada and Russia. The gene map for the American mink (Serov & Pack 1993) is not yet
well developed compared to other species of domestic animals. However Zoo-FISH mapping for
the mink has been carried out (Hameister et al. 1997), showing that 34 human chromosome
segments cover the entire mink genome and have syntenic relationship with human
chromosomes.
In human there have been several reports on the frequency of disomy on a number of individual
chromosomes. The frequencies of disomy for normal males has been estimated to 0.2-0.3 per
cent for each chromosome (Pellestor et al. 1996). Disomy frequencies has been used to the
estimate effects of environmental hazards or diseases on the production of abnormal sperms in
human males (Monteil et al. 1997 and Robbins et al. 1997).
The present report deals with mink cosmids with repeats which showed strong FISH signal on
interphase cells. The cosmids are described by Christensen et al. (1997), and chromosome
specific cosmids were available for the mink chromosomes No 8, 9, 11 and Y. Another cosmid
containing nuclear organizer sequences hybridize to chromosome 2 and 5. The five probes has
been used individually on sperms from normal males mink to evaluate the frequency of disomy.
Material and methods
The animals: The mink were from a normal production line of the Wild mink or Pastel colour type. They were kept under normal farming condition and sperm were collected from the epididymus at the end of the mating season at the 24 Marts and at the 1st April after pelting.
Sperm preparation: After collection the semen was washed 3 times in PBS and finally stored
in 70% ethanol at 5oC. The sperm were dropped on a wet slide using the fixative (methanol-acetic
acid 3:1) and washed with two drops of fixative put on the top of the tilted slide, the fixative were
sucked of at the edges and the rest was air dried. The slides were kept for one week at room
temperature. Finally the slides were treated with dithiotreitol (DTT) (200 mM KOH, 50 mM
DTT) under a cover slip, pulling the cover slip over the slide to give a varying exposure time of
.5-2 min. The cover slips were washed of with distill water and the slide were washed in the
neutralizing buffer (900mM Tris-HCl, pH 8.3, 300mM KCl, 200 mM HCl) and finally washed
with distil water and air dried.
DNA probes: Five probes, AG25 (chromosome Y), AG32 (chromosome 11), AG34 (chromosome
2 and 5), AG63 (chromosome 9) and AG64 (chromosome 8) were double labelled with nick
translation with biotin-14-dCTP and biotin-14-dATP.
FISH: Denaturation of the DNA in the sperms were done with 70 % formamide/2xSSC, pH 7.0,
at 65oC for 2 min and thereafter washed in an ethanol series on ice 70% 90% 100% each for 2
min..There was used one µg biotinylated probe in 30 µl of hybridization solution containing 45
% formamide, 2 x SSC, 10% dextran sulfate. The hybridization mix was supplemented with 6
µg sheared genomic mink DNA; denaturation at 70oC for 5 min and incubated for 20 min. at
37oC.In situ hybridization was carried out by incubating the slides with denatured sperms and
hybridization mix at 42oC for 20 hours. After hybridization, the slides were washed two times in
45% formamide, and three times in 2 x SSC at 39-42oC. Visualization of the biotinylated probe
was achieved with fluorescein isothiocyanate (FITC) conjugated to avidin, and the signal was
amplified with one layer of biotinylated anti-avidin antibody (Vector Laboratories). For more
details see Thomsen et al. (1996).
Microscopic observations: Only well delineated sperm cells were counted and double spots were
scored when an extra signal spot occurred being of the same size as the normal ones, and the two
spots separated by at least one spot diameter.
Results and discussion
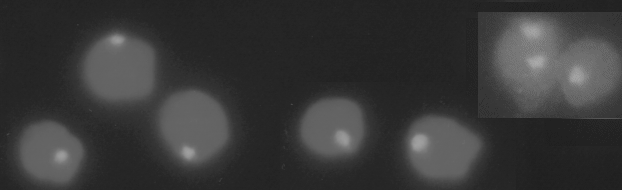
In Fig. 1 can be seen sperm cells with the strong hybridization signal, the probe for chromosome
8 gives very well demarked spots. More details are given by Christensen ( 1997).
Fig. 1. Hybridization signals form the cosmids AG64 (chromosome 8) on mink sperm cells.
Table 1. Preliminary counts of disomy in Mink sperm cells for chromosome 8 and 9 and 11
| Animal | Probe | Normal | Disomic | No signal |
| 9236 | AG64 (chr 8) | 2340 | 12 | 58 |
| 9238 | AG63 (chr 9) | 2040 | 11 | 93 |
| 9238 | AG32 (chr 11) | 1127 | 3 | 55 |
In Table 1 are given the disomy counting results for two animals with a probe for chromosome
8, 9 and 11. The disomy rates for the three chromosomes are estimated to 0,51, 0,54 and 0,27 per
cent, respectively.
Until now very few strong centromere repeat probes has been developed for the domestic animal species covering single chromosomes. For the domestic pig there exist a chromosome specific strong repeat probe for chromosome 1 (Jantsch et al. 1990) and the Y chromosome (McGraw et al. 1988), which have been used for estimating the frequency of Y bearing sperms (Kawarasaki et al. 1996). In human there has been developed -satellite centromere probes for almost every chromosome and they have been used to estimate the rate of disomy in both normal and abnormal human males.
There have been some difficulties with the evaluation of disomy. The probe for chromosome 8 gives very distinct spots as can be seen in Figure 1 whereas the probes for 9 and 11 give more long and narrow spots which might be interrupted with no signal. Until now we were not able to set the conditions for counting the signal from the nuclear organizer probe, chromosome 2 and 5, as the two chromosomes often lie close together and the signal fuse into one. The use of the Y chromosome probe also needs refinements before data can be given.
The frequency of disomy per chromosome in humans range in the neighbourhood of 0.2 per cent
In this study in the mink the frequency are around the double. To study the frequency of diploid
sperms it is necessary to use two probes with multicolour FISH. In this study only one probe has
been used per experiment, so it has not been possible to separate disomic and diploid sperms
which might overestimate the rate of disomy. With these reservations the results for the mink
here given should be taken as preliminary, more cells will be counted and the technique refined.
The technique is worth while to develop for all other domestic animals species. It should be a
mean to understand and thereby improve aspects of fertility.
Acknowledgement. Thank to Inger H. Christensen for excellent technical assistance. The work
was supported by the Ministry of Agriculture.
References:
Christensen, K. (1997) http://www.husdyr.kvl.dk/htm/kc/mink/sperma/sperma.htm.
Christensen, K., Brusgaard, K., Malchenko, S., Lohi, O., and Serov, O., (1997) Arch. Zootec 45:259-265.
Hameister, H., Klett, C., Bruch, J., Vogel, W. And Christensen, K., (1997). Chromosome Research, 5:5-11.
Jantsch, M., Hamilton, B., Mayr, B. & Schweizer, D. (1990) Chromosoma (Berl) 99:330-335.
Kawarasaki T., Sone M., Yoshida M., and Bamba K. (1996) Mol. Reprod.Dev. 43: 548-553.
McGraw, R.A., Jacobson, R.J. & Akamatsu, M. 1988. Nucl. Acids Res.16:10389.
Monteil, M., Rousseaux, S., Chervet, E., Pelletier, R., Cozzi, J, and Sele, B., (1997). Cytogenet. Cell Genet. 76:134-138.
Pellestor, F., Girardet, A., Coignet, L., Andreo, B., and Charlieu J.P., (1996). Am. J. Hum. Genet. 58:797-802.
Robbins, W.A., Meistrich, M.L., Moore, D., Hagemeister, F., Weier, H.-.W, Cassel, M.J., Wilson, G., Eskenazi, B.and Wyrobek, A.J., (1997) Nature Genet. 16:74-78.
Serov, O.L. and Pack S.D., (1993) In Genetic Maps, ed. SJ O'Brian. 279-281.
Thomsen, P.D., Høyheim, B. and Christensen, K., (1996). Cytogenet. Cell Genet.73:203-208.