Breed the right animals. Are you in doubt? Use paternity testing.
By K. Christensen, R. Anistoroaei and A. Farid
The Royal Vet. & Agric University, Department of Animal Science and Animal Health, Genetics and Breeding, Grønnegårdsvej 3, DK-1870 Frederiksberg C, Denmark
Paternity testing in mink by means of DNA technological methods is now available. The test can be carried out on a capillary tube blood sample or a small hair sample (around 100 hairs including under fur hair containing the roots plucked from the tail). The test can be done in a week and it can solve the uncertainty of paternity of your mink. The test can typically answer the question: can this male or female be sire/dam of this litter/animal? Or do this animal belong to a group of full sibling?
The DNA markers making the paternity testing possible has been developed by participating in an International Mink Gene Mapping Project. Furthermore, the goal of this project is to assign important mink genes to specific chromosome location so they can be followed by genetic markers.
The traditional paternity test has been done by means of blod types in man and the large domestic during the last 50 years. But today it is well known that any doubt of the identity of a person can be resolved by means of the modern DNA technological methods. In the mink there has never been developed any of the old methods for identifying an animal. This has been due to the comparatively low reproduction rate found in the mink and there by the relatively little significance of the individual animal. But with the development of the DNA technological methods paternity testing has been adobted as a check on the pedigrees and in some cases also for resolving the paternity in dogs and cats. Typically if two sires have mated a female in the same oestrus the question is which one is the sire of a specific pup in the resulting litter. Here we will describe shortly how to do to resolve a disputed pedigree or in identification of carrier also in the mink.
The samples
The paternity test requires samples from the mother, a potential father, and the offspring. If the mother is not available, testing can still be performed using more extensive procedures based on full an half sib.
The sample can be:
1. tail hair pulled out so the hair roots come withe the hair,
2. a nail clip containing a little blod or
3. capillary tube blood
The sample should imediately be placed in a sterile plastic bag or tube labelled with the animal identity. The collecting of the samples must be as sterile and accurate as possible. The samples can be posted and it should reach the laboratory within two days. A draft or a list over the expected relationship between the animals should follow the samples.
The methods
The laboratory methods are based on DNA polymorphisms for 'Short Tandem Repeat' the so-called mikrosatellit markers. This type of polymorphism is available in large quantities in all animal species, and at the moment up to 100 different systems are ready for use in the mink, (Brusgaard et al 1998 and Vincent et al. 2003). An accurate paternity verification would normally require investigation of 10 to 15 systems. The typing is based on the 'Polymerase Chain Reaction' (PCR) technology. This technique is based upon the natural amplification process of DNA. By means of the PCR technology the mothers and fathers alleles can be made visible in the offspring after electrophoresis. An accurate report is based on the combined results of at least 10 to systems.
The Genetics
The genetic rules are very simple. An individual can maximally carry two alleles in a marker system. One received from the father and the other received from the mother. If an allel in a presumed offspring is different from the ones occurring in the parents, the parentage can be excluded for a parent not contributing an allel. Opposite, if 10 marker systems tested for a presumed offspring adhere to the rule the paternity can be approved.
The paternity test
In Figure 1 is shown how the alleles are made visible on a computer screen after electrophoresis.
Figure 1. Illustrate result of running two DNA marker systems in a family consisting of father, mother and 4 offspring. The offspring are suspected to have been mixed up. The offspring 1, 3 and 4 show only peaks found in either the father or the mother. Whereas the markers of 'offspring' number 2 clearly show that it does not belong to the sib ship, none of its markers can be found in fathers and mothers lane. In table 1 is given an interpretation of the peaks from this figure.
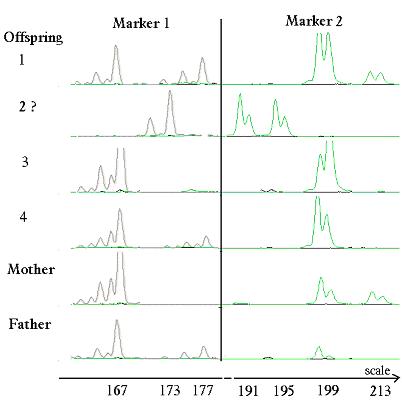
Table 1. Interpretation of DNA markers shown in Figure 1, where to each allele is given a number corresponding to its place on the scale. An animal can only have two alleles in each marker system (Mvi1272 and Mvi1273 Vincent et al. 2003), one received from its father and one from its mother. Only the major peaks, can be interpreted as alleles. The mother has only one allele in marker 2 and the father only one allele in marker 1. In such cases they carry two identical alleles and they are designated as homozygotes.
Marker 1 (Mvi1273) Marker 2 (Mvi1272) Offspring 1 199/213 167/177 Offspring 2? 191/195 173/173 Offspring 3 199/199 167/167 Offspring 4 199/199 167/177 Mother 199/199 167/177 Father 199/213 167/167
The test should be based on at least 10 marker systems. In this example are shown the results of two of the systems, only, namely those where offspring number two deviate from the expected alleles it should have received from its parents. A rejection of the correct paternity should be based on deviations in at least two marker systems to exclude technical errors causing the deviation. To certify the paternity, all systems should confer to the rule that there aren't any alleles in the offspring, which are not present in the parents.
From the genetic rules and results given in Figure 1, non-affiliation of offspring 2 to the parents is obvious.
The Section of Animal Genetics at the Royal Vet. & Agric. University, Frederiksberg, Denmark can from now on provide you with paternity testing in mink by means of DNA technological methods provided that the expenditures should be covered.
Literature
Brusgaard, K., Shurki, N., Malchenko, S., Lohi, O., Christensen, K. & Kruse, T.A. (1998) Three polymorphic mink, Mustela vison, dinucleotide repeats. Animal Genetics, 29, 153-153.
Vincent, I.R., Farid, A. and Otieno. C.J. (2003). Variability of thirteen microsatellite markers in American mink (Mustela vison). Canadian Journal of animal Genetics 83:597-599.
Acknowledgements
The Danish Fur Breeders Association has supported this project.