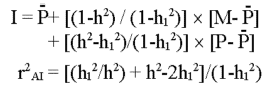
For large domestic animal breeding organizations estimation of breeding values is an enterprise which can
give the members a large economic return. Therefore, very refined methods are
applied to improve the index estimation,
also see lections 12.1-2. In addition to what is shown here in terms of simple
estimation of breeding
value, it is of great importance to corrected for significant environmental factors, as for
instance seasons and herd average. It is also important
that all related individuals
contribute with information to the indexes.
Estimation of the breeding value is based on large equation systems for all animals in the
population simultaneously. This provides the possibility of simultaneous correction
of environmental effects.
Estimation of the breeding value of an animal can be calculated based on all possible combinations of information. Section 7.2 only shows examples in which uniform relationship exists for the phenotype measurements. Her only a few exceptions from this form shall be shown. The first one is estimation of the breeding value based on the estimates of the parents' breeding value, which is which is also shown in section 7.3:
Ioffspring = (Isire + Idam)/2 and r2AI,offspring = (r2AI,sire + r2AI,dam)/4
Below is shown by formulas to calculate how much a gene marker can add to the information from a phenotype measurement. The trait's
heritability is h2 and the gene marker represents a part of the joint heritability,
which corresponds to (h1)2.
When M is the marker effect and P the phenotype value, the estimated breeding value
(I) is as follows:
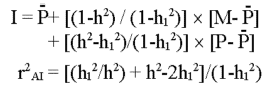
In the table below are results from the use of the formula for accuracy with and without a gene marker (large effect, 20% of the genetic variation, column +M).
-M +M -M +M -M +M Transferrin locus -------------------------------------------------------------------------- h2 .05 .05 .25 .25 .50 .50 | .33 (h1)2 0 .01 0 .05 0 .10 | .01 rAI2 .05 .24 .25 .35 .50 .56 | .34
It appears that, for traits with low heritability, a significant improvement of the accuracy can be obtained by using the information on the gene marker. For traits with high heritability the improvement is very low.
Example: Index of a cow with a yield of 2300 kg milk and Tt in transferrin type from the population described in section 6.2. This cow becomes like the marker genotype (M=1882 kg milk) and its mean value is (Pave.=1904).
The formula above is applied and h2 and h12 are taken from the table
I = Pave + X*(M - Pbar ) + Y*( P - Pbar ),where X = (1-h2)/(1-h12) = (1 - 0,33)/(1 - 0,01) = 0,68 Y = (h2-h12)/(1-h12) = (0,33 - 0,01)/(1 - 0,01) = 0,32 by insertion is obtained I = 1904 + 0,68*(1882-1904) + 0,32*(2300-1904) = 2016 kg milk
The accuracy as is seen from the table is only slightly improved by using the transferrin type in the index.
For estimating the effect of a gene influencing a quantitative trait (a QTL), reference can be made to lection 12.5.
Standardization of a breeding value index.
The breeding value indexes are normally expressed in absolute units. The index,
for instance, of a bull with a daily weight gain of 1100 grams from birth
to 12 months, many people have difficulties in deciding whether this is good or bad. The
basis for the comparison is missing when the mean value is unknown.
Therefore, by subtraction of the mean value the indexes are centered around zero.
Thus the better half will have a positive index and the worse half will have a
negative index.
Furthermore the indexes can be standardized both by subtracting the mean value and dividing
it with its standard deviation. Such a population will have a mean
value of zero and a standard deviation of one.
In Denmark standardized indexes are often used, so the mean value is 100 and
a standard deviation of for instance 5. With such an
index animals with values higher than 100 are better
than the average and animals with an index
value lower than 100 are worse than the average.