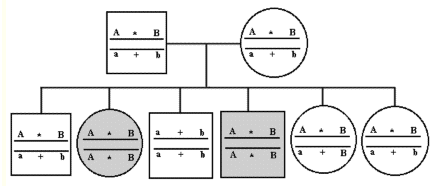
Genealogical graph of gamete types of three linked loci. The gamete types are deduced from lab tests of locus A and B and the segregation of the offspring. The disease gene based on the two diseased offspring.
Use of DNA markers within families
If segregation of a recessive disease occurs within a family and if information of the localization on
the chromosome exists and if flanking markers is known, it is possible to identify carriers of the gene, even if the
genetic code of the gene is not known.
Method: Figure 12.1 shows segregation of markers A and B, as well as a disease gene (*). The two diseased animals in the figure are full sibs. All the sibs and the parents have been examined for alleles in locus A and B. Among the offspring the two diseased ones have the genotypes AABB. Based on the offspring the haplotype of the parents can now be interfered. The gametes AB must be carriers of the disease gene due to linkage.
| Figure 12.1. Genealogical graph of gamete types of three linked loci. The gamete types are deduced from lab tests of locus A and B and the segregation of the offspring. The disease gene based on the two diseased offspring. |

|
The fifth offspring has a recombination of the gene marker B and the disease gene,
since is genotype AaBB
and not affected.
In many cases only a single DNA marker exists, as not all domestic animal
species can have that many markers. For the DNA marker to have any value it is
necessary that the parents are heterozygotic.
Summary of the investigation: Animals with the genotype aabb do not carry the disease gene. Whereas all genotypes
with at least one allele carrying a capital letter are expected to be carriers of the disease gene.
By moving backwards in the
pedigree, it is evident that all individuals with at least one allele with a capital
letter are possible carriers of the disease
gene.
DNA markers applied on population level
For a DNA marker to be applied on population level it has to be based on linkage disequilibrium and very close
linkage with the disease gene. Strong linkage disequilibrium always exits if the mutation has occurred
within a
few generations back. A substantial amount of linkage disequilibrium occur even after 10-15 generations,
if the distance to the marker is less than 1 cM, see section 2.5. Use of a marker on population level would only
indicate a possible carrier, which has a higher chance of being carrier than a randomly chosen animal. The final
proof can only be established by segregation of diseased individuals.
To conclude this section on anonymous DNA markers it needs to be said, that use of a DNA marker for a disease gene is only second best. The goal should always be to find the mutation and to examine the DNA type to find the real culprit. Though the DNA markers are still to be used.